© 2023 Melvil Lardinois
741 New South Head Rd, Triple Bay SWFW 3148, New York
Data science, Data model, Tech advisor

2020 - 2022
In 2020, I got in contact with the data project manager at ICO through my network. At the time ICO just signed a deal with IQVIA, the biggest American healthcare data company, with the objective to build a structured data warehouse to participate in clinical trials.
Official websites :
ICO
IQVIA
I was invited to the kick-off meetings as a tech advisor for ICO. It was the first time that the institute was entering the domain of data science and software development, therefore they needed guidance to understand and challenge the demands of IQVIA, to size each task, and to make sure that they weren’t being sold unadapted tools.
Initially, I was just giving free consulting because I was very interested in the project, but IQVIA was struggling to find developers in France to work on the project so I ended up joining the ICO tech team (the Data Factory) officially with one of my team member a few months later.
This was my first time working in the Healthcare industry so my priority was to learn and get used to the medical vocabulary quickly. I moved back to France to be able to go to the hospital in person and talk to heads of departments, doctors, and patients to gather as much information as possible.
Once I got an overall understanding of how things worked there and the kind of company culture I was stepping in, I started analysing which tech solutions would suit my client's needs better taking into consideration the budget and the existing infrastructure. It took a while to figure out the big picture as each medical service has its own system and equally complex concepts but it was a necessary effort.
I was positively welcomed by everyone involved in the project. People were eager to help and demanding for new ways to investigate their patient’s data, as long as we are protecting them. The main thing to keep in mind is that doctors’ calendars are fully booked as they are busy saving people’s lives, so you cannot waste their time.
For confidentiality reasons I can’t get too in-depth with the examples, but I will try to give a comprehensive explanation of our mission and how we approached the different issues.
Patients' data are extremely valuable for expanding analysis and finding patterns between pathologies and treatments. The major issue is that these data are located in consultation reports, spread into different systems, and in different formats. So we need to find ways to transform this unstructured data into structured and standardized tables. We had a limited budget so we tried to reuse our code and methodology as much as possible.
The first step was to identify the most important “variables of interest” and assess them one by one to figure out where we could find these data and how to extract them from the data source. There are literally hundreds of them so we prioritized based on the easy ones to get and the most requested ones as well. During this process, we also spent a lot of time talking to other hospitals and 3rd party healthcare data solution providers to validate our approach and figure out if we should purchase proprietary software to progress quicker. Generally, the bottom line is that is it cheaper and more efficient to develop our own code and pipeline because you still need to allocate resources to set up a new software and each hospital data system is so different that you end up having to build a lot of custom code anyway.
Example of structured data (ST) : birthdate, sex, pathology, treatment_name, treatment_startdate
Example of unstructured data (NST) : death_cause, tumour_histological_type, metastatic_locations, treatment_evolution
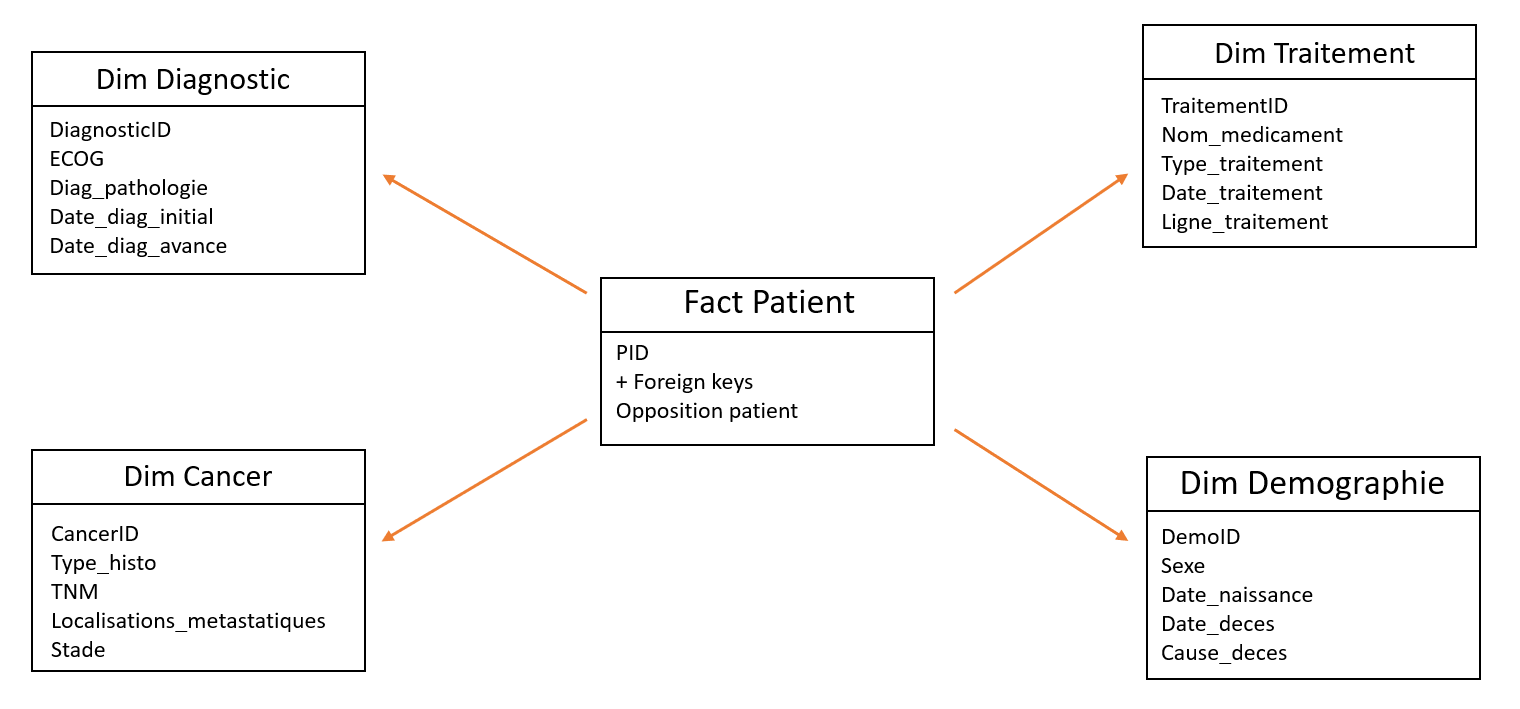
1st iteration of the data warehouse star schema
When looking to extract variables of interest, there are 4 cases possible:
More details on these processes below.
Context
The medical staff fills in a survey about the patient’s medical history (smoking, alcoholism, comorbidity...). These answers are in free text (see example), which makes it almost impossible to process this data without reorganizing it first. We want to categorize these responses and extract specific data that could be relevant for further analysis. This is a relatively simple task that could be done by a human. The problem is that there are tens of thousands of data points to process and it would be time consuming, expensive and inefficient to do this work by hand. Our goal is to do this automatically using machine learning algorithms. In this part, I will focus on the NLP process and will go more in-depth about regex extraction in the following section.
Example
Question: Are you a smoker?
Answer: Smoking brake for about 6 years, estimated at about 15 packs per year.
Expected result
Categories
0- Non smoker
1- Smoker
2- Former smoker
Data extraction
Nb of packs per year: 15
Weaned for: 6 years
Solutions & technical explanations
• Libraries and algorithms
To speed up much of our work, we use open source libraries developed by other computer developers. A library is a set of algorithms that revolve around a single theme. For example, Pandas is a Python library that includes many mathematical functions for data analysis and visualization. For the classification problem, we were particularly interested in Natural Language Processing (NLP) libraries.
• Process details
NLP is one of the applications of machine learning. Specialized in the understanding and processing of human language, the idea is to train NLP algorithms to correctly classify patients' free text responses. To do this, we labeled a few hundred pieces of data by hand, i.e. we extracted the data from a data warehouse and assigned the correct category to each of these answers. This is a tedious but necessary process to provide content for our NLP classification model. Finally, we test the trained models to check the accuracy of the results obtained.
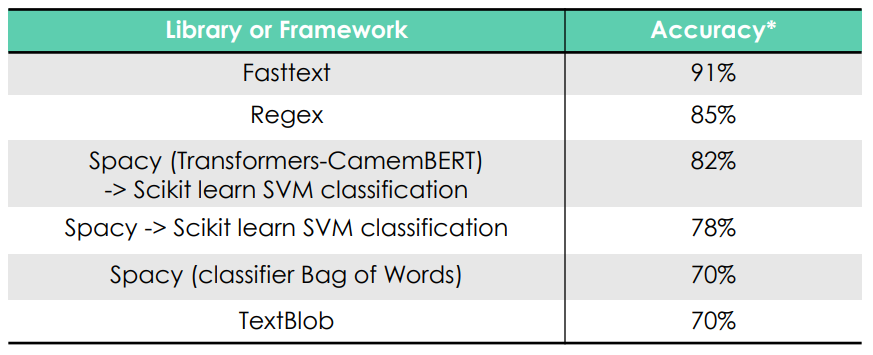
Library comparison (*for a train set of 100 records and a test set of 100 records)
Fasttext had the best results so we kept using this library for the rest of the project. The data set was extracted randomly from our data source. We have labeled 200 more data to grow our train and test sets (now 400 total).
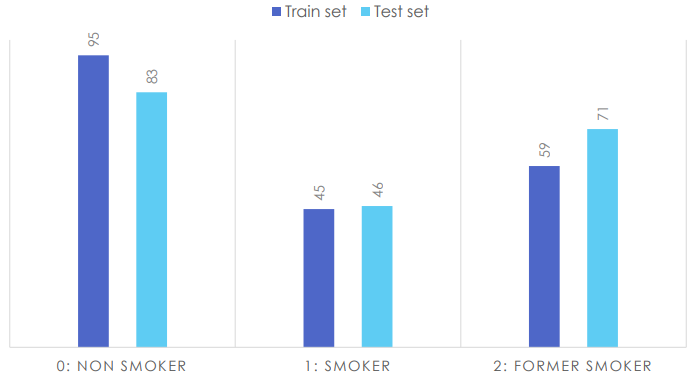
Dataset distribution
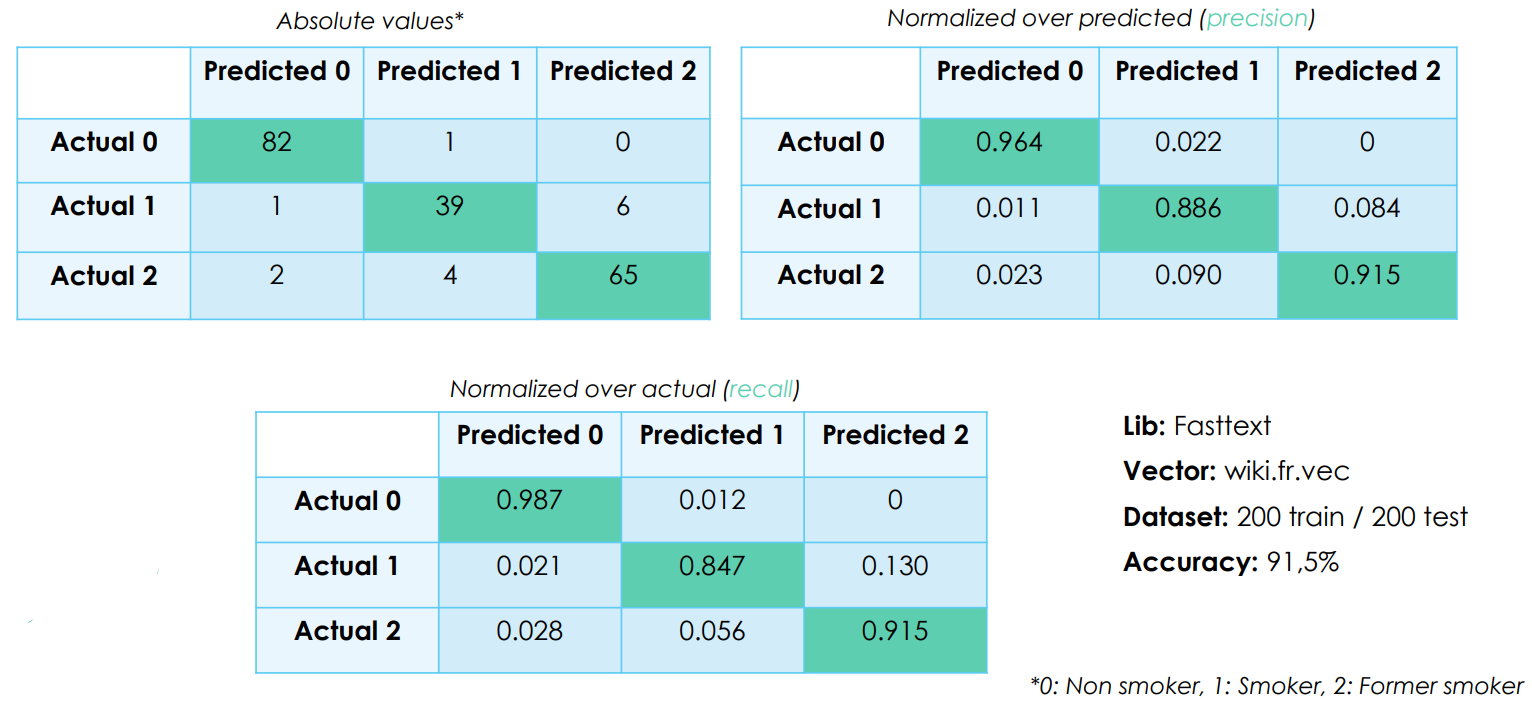
Confusion matrix
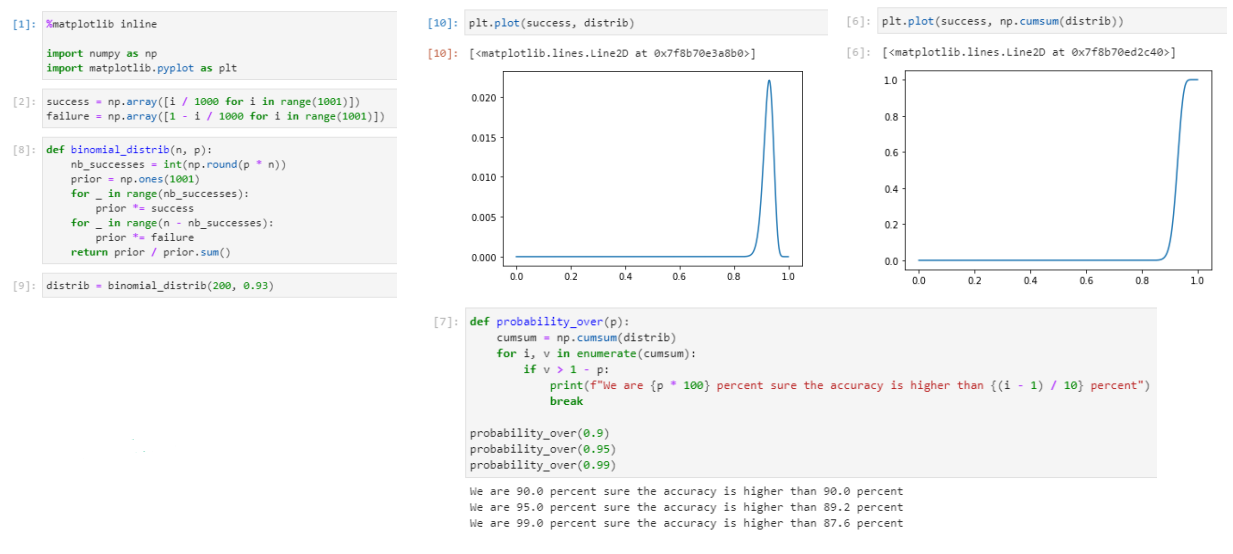
Binomial distribution & Code sample
We achieved a 91,5% accuracy, a number that we double checked thanks to the binomial distribution. It's good, but it could be improved.
During categorization, the NLP model will always categorize the data, even when it is not sure of itself at all. Sometimes it will be 99% sure that its categorization is the right one, sometimes it will only be 30% sure. If we accept not taking into account the results that are below the 50% certainty threshold, for example, which will cause us to "lose" some of the data in the entire dataset, we could increase the accuracy of the model on all the remaining data.
When the source documents are exploitable in a text or PDF format, we have to find a way to extract words or numerical values that belongs to the list of variables of interest.
The most common practice is to use regex. A regular expression (shortened as regex) is a sequence of characters that specifies a search pattern in a text. Usually, such patterns are used by string-searching algorithms for "find" or "find and replace" operations on strings, or for input validation.
Here is an example :
We want to extract any variable that can give us information about the karnofsky performance status. Usually, it’s a number between 0 and 100 that represents the patient’s general well-being and activities of daily life. So we’re scanning for words like “ecog”, “oms”, “ps, “performance status”, ...
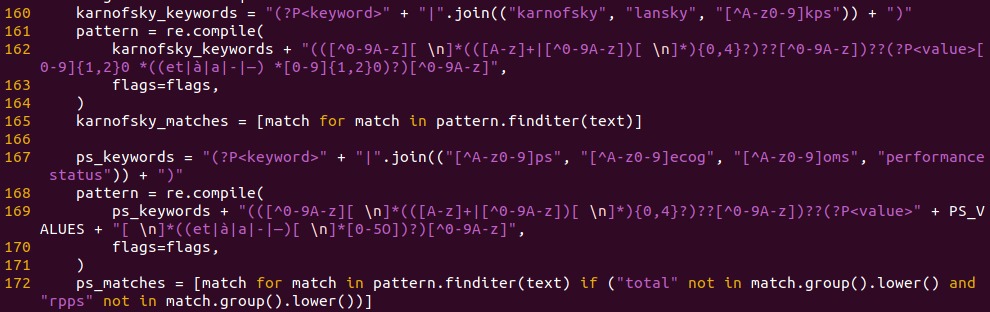
Regex example
We made a simple web interface to visualize the results more easily.

Regex result
We still have to homogenize the results because some of the performance status are expressed on a 1-5 scale, and others with a 0-100 scale (it depends on the doctor writing the report). Finally, we can insert these values into our data warehouse.
A better option than using regex would be to train an NLP (natural language processing) algorithm to understand these documents so we wouldn’t have to make a list of keywords for each variable of interest. Comprehend Medical, an AWS entity recognition solution, is particularly good at this but isn't available in French yet.
In many cases, the data we are interested in is located inside documents that have been scanned, especially when they come from external medical facilities. Therefore, this content can’t be accessed until it is transformed into selectable text. The solution is to use an OCR (optical character recognition) algorithm that is as accurate as possible to generate a perfect copy of the initial document, but with selectable text this time.
In our research, we tried to build our own OCR solution using open source libraries and improving it with custom code. Here is an example of the best output we could achieve in a week of work :


It’s pretty good, but it is absolutely necessary to have 99% - 100% accuracy because if any numerical value is wrong, the results are completely unusable. Therefore, we started looking at the proprietary solutions from different cloud providers. The conclusion was that AWS Textract, the OCR solution from AWS, was miles ahead of what we could ever develop ourselves.
While it is not optimal for us to use a cloud solution because we have to anonymize all the data that is sent to the cloud and the client has to pay for the service, it’s still the only option because the incredible performances and quality of their algorithm match our business requirements.
With the overall aim of developing methodologies and open-source code that can be extended and applied to other medical institutes, we wrote a scientific paper about one of the use cases we worked on.
We published this paper titled "Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France" in the magazine " IJERPH" for the special issue "The Revolution of Health Data Warehouses in Hospitals: From Theory to Practice".
Abstract
Electronic Medical Records (EMR) and Electronic Health Records (EHR) are often missing critical information about the death of a patient, although it is an essential metric for medical research in oncology to assess survival outcomes, particularly for evaluating the efficacy of new therapeutic approaches. We used open government data in France from 1970 to September 2021 to identify deceased patients and match them with patient data collected from the Institut de Cancérologie de l'Ouest (ICO) data warehouse (Integrated Center of Oncology-the third largest cancer center in France) between January 2015 and November 2021. To meet our objective, we evaluated algorithms to perform a deterministic record linkage: an exact matching algorithm and a fuzzy matching algorithm. Because we lacked reference data, we needed to assess the algorithms by estimating the number of homonyms that could lead to false links, using the same open dataset of deceased persons in France. The exact matching algorithm allowed us to double the number of dates of death in the ICO data warehouse, and the fuzzy matching algorithm tripled it. Studying homonyms assured us that there was a low risk of misidentification, with precision values of 99.96% for the exact matching and 99.68% for the fuzzy matching. However, estimating the number of false negatives proved more difficult than anticipated. Nevertheless, using open government data can be a highly interesting way to improve the completeness of the date of death variable for oncology patients in data warehouses.
You can find the full paper here.
Everything was going well until ICO hired a new project manager for the Data Factory. This person failed to understand and adapt to the company, making too many sub optimal decisions, partnering with the wrong third parties, and wasn’t able to prioritize the team’s tasks properly. This led to a slow breakdown of the project until it wasn’t possible to work on anything at all.
Thus we ended up quitting in May 2022. While I’m glad about the opportunity we had and all the things we could learn along the way, I’m personally very disappointed how our relationship shaded and I wish that one day we can come back in different circumstances and finish what we started.